Sitemap
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Pages
Bioinformatics Workshop Projects
Study projects completed as part of the workshops undertaken during the training at the Bioinformatics Institute:
1. Variant calling of Escherichia coli WGS
2. Variant calling of deep sequencing data (Influenza A virus (H3N2) hemagglutinin gene)
3. De novo assembly of Escherichia coli genome
4. Tardigrade Ramazzottius varieornatus genome annotation and protein function prediction
5. Genotyping and SNP annotation of human 23andMe data
6. RNA-seq data analysis for differential gene expression of Saccharomyces cerevisiae after 30 minutes of fermentation
7. Ancient metagenomes analysis examining human dental calculus
8. Annotation of the immune repertoire derived from the T-cell population in a relatively healthy donor
9. Single-cell CITE-seq analysis detailing the cellular composition and transcriptional profiles within human bone marrow
SequenceForge-Lite
About:
Status:
About: Lightweight tool designed to work with biological sequence data, providing various functionalities for filtering FASTQ files and manipulating FASTA files
Status: Pre-release. In seek of ideas how to make it perfect
Posts
publications
Morphogenesis of penile cavernous fibrosis in hypotestosteronemia: an experimental study
Kogan M.I., Todorov S.S., Popov I.V., Popov I.V., Kulishova M.A., Ermakov A.M., Sizyakin D.V.
Urology Herald, 2020
Visualisation of penile structures of laboratory rabbit: ultrasound, histology, and micro-CT
Kogan M., Popov I., Mitrin B., Popov I., Sadyrin E., Pasetchnik D., Ermakov A., Ugrekhelidze N., Kulikova N.
E3S Web of Conferences, 2020
Coronavirus infections of animals: Future risks to humans
Donnik I.M., Popov Ig.V., Sereda S.V., Popov Il.V., Chikindas M.L., Ermakov A.M.
Biology Bulletin of the Russian Academy of Sciences, 2021
X‐ray micro‐computed tomography in the assessment of penile cavernous fibrosis in a rabbit castration model
Kogan M.I., Popov I.V., Kirichenko E.Y., Mitrin B.I., Sadyrin E.V., Kulaeva E.D., Popov I.V., Kulba S.N., Logvinov A.K., Akimenko M.A., Pasechnik D.G., Tkachev S.Yu., Karnaukhov N.S., Lapteva T.O., Sukhar I.A., Maksimov A.Yu., Ermakov A.M.
Andrology, 2021
Clinical cases of infectious endocarditis in cats
Sereda T.V., Petrova M.A., Popov I.V., Popov I.V., Kartashov S.N., Ermakov A.M.
Brazilian Journal of Veterinary Medicine, 2022
Coronaviruses of synantropic bats: an unexplored threat
Lipilkina T., Popov I., Kitsenko K., Popov I., Ermakov A.
E3S Web of Conferences, 2022
Detection of coronaviruses in insectivorous bats of Fore-Caucasus, 2021
Popov I.V., Ohlopkova O.V., Donnik I.M., Zolotukhin P.V., Umanets A., Golovin S.N., Malinovkin A.V., Belanova A.A., Lipilkin P.V., Lipilkina T.A., Popov I.V., Logvinov A.K., Dubovitsky N.A., Stolbunova K.A., Sobolev I.A., Alekseev A.Yu., Shestopalov A.M., Burkova V.N., Chikindas M.L., Venema K., Ermakov A.M.
Scientific Reports, 2023
Cultivable Gut Microbiota in Synanthropic Bats: Shifts of Its Composition and Diversity Associated with Hibernation
Popov I.V., Berezinskaia I.S., Popov I.V., Martiusheva I.B., Tkacheva E.V., Gorobets V.E., Tikhmeneva I.A., Aleshukina A.V., Tverdokhlebova T.I., Chikindas M.L., Venema K., Ermakov A.M.
Animals, 2023
Gut Microbiota Composition of Insectivorous Synanthropic and Fructivorous Zoo Bats: A Direct Metagenomic Comparison
Popov I.V., Popov I.V., Krikunova A.A., Lipilkina T.A., Derezina T.N., Chikindas M.L., Venema K., Ermakov A.M.
International Journal of Molecular Sciences, 2023
Gut microbiota differences in stunted and normal-length children aged 36–45 months in East Nusa Tenggara, Indonesia
Surono I.S., Popov I., Verbruggen S., Verhoeven J., Kusumo P.D., Venema K.
PloS ONE, 2024
Effects of spore-forming Bacillus probiotics on growth performance, intestinal morphology, and immune system of broilers housed on deep litter
Popov I.V., Skripkin V.S., Mazanko M.S., Epimakhova E.E., Prazdnova E.V., Dilekova O.V., Dannikov S.P., Popov I.V., Trukhachev V.I., Rastovarov E.I., Derezina T.N., Kochetkova N.A., Weeks R.M., Ermakov A.M., Chikindas M.L.
Journal of Applied Poultry Research, 2024
Detection of Brno loanvirus (Loanvirus brunaense) in common noctule bats (Nyctalus noctula) in Southern Russia
Ohlopkova O.V., Stolbunova K.A., Popov I.V., Popov I.V., Kabwe E., Davidyuk Yu.N., Stepanyuk M.A., Moshkin A.D., Kononova Yu.V., Lukbanova E.A., Ermakov A.M., Chikindas M.L., Sobolev I.A., Khaiboullina S.F., Shestopalov A.M.
Brazilian Journal of Microbiology, 2024
Detection and Phylogenetic Analysis of Alphacoronaviruses in Bat Populations of Rostov and Novosibirsk Regions of Russia, 2021–2023
Ohlopkova, O.V., Popov, I.V., Popov, I.V., Stolbunova, K.A., Stepanyuk, M.A., Moshkin, A.D., Maslov, A.A., Sobolev, I.A., Malinovkin, A.V., Tkacheva, E.V., Bogdanova, D.A., Lukbanova, E.A., Ermakov, A.M., Alekseev, A.A., Todorov, S.D., Shestopalov, A.M.
Microbiology Research, 2024
Differences in gut microbiota composition, diversity, and predicted functional activity between wild and captive zoo Carollia perspicillata in a One Health perspective
Popov I.V., Popov I.V., Chebotareva I.P., Tikhmeneva I.A., Peshkova D.A., Krikunova A.A., Tkacheva E.V., Algburi A.R., Abdulhameed A.M., Jargalsaikhan A., Ganbold O., Chikindas M.L., Venema K., Ermakov A.M.
Brazilian Journal of Microbiology, 2025
Identification of orthohantaviruses detected for the first time in the Republic of Belarus
Semizhon P.A., Scheslenok E.P., Dubkov N.A., Sukhotskaya E.A., Stolbunova K.A., Popov I.V., Popov I.V., Alekseev A.Y., Kabwe E., Davidyuk Y.N.
Problems of Virology, 2025
Gut Microbiota Dynamics in Hibernating and Active Nyctalus noctula: Hibernation-Associated Loss of Diversity and Anaerobe Enrichment
Popov I.V., Peshkova D.A., Lukbanova E.A., Tsurkova I.S., Emelyantsev S.A., Krikunova A.A., Malinovkin A.V., Chikindas M.L., Ermakov A.M., Popov I.V.
Veterinary Sciences, 2025
Metagenomic Investigation of Intestinal Microbiota of Insectivorous Synanthropic Bats: Densoviruses, Antibiotic-Resistance Genes, and Functional Profiling of Gut Microbial Communities
Popov I.V., Manakhov A.D., Gorobets V.E., Diakova K.B., Lukbanova E.A., Malinovkin A.V., Venema K., Ermakov A.M., Popov I.V.
International Journal of Molecular Sciences, 2025
Genomic insights into Streptomyces albidoflavus SM254: tracing the putative signs of anti-Pseudogymnoascus destructans properties
Popov I.V., Chikindas M.L., Popov I.V.
Brazilian Journal of Microbiology, 2025
Beyond White-Nose Syndrome: Mitochondrial and Functional Genomics of Pseudogymnoascus destructans
Popov I.V., Todorov S.D., Chikindas M.L., Venema K., Ermakov A.M., Popov I.V.
Journal of Fungi, 2025
Beneficial Properties of Enterococcus faecium Strains Isolated From Bats’ Fecal Samples
Lipilkina T.A., Alves M.V., Carneiro K.O., Leani K., Popov I.V., Popov I.V., Chikindas M.L., Ermakov A.M., Todorov S.D.
Molecular Nutrition & Food Research, 2025
KEGGaNOG: A Lightweight Tool for KEGG Module Profiling From Orthology-Based Annotations
Popov I.V., Chikindas M.L., Venema K., Ermakov A.M., Popov I.V.
Molecular Nutrition & Food Research, 2025
research
Pediatric Nutritional Medicine and Intestinal Microbiology
Research Internship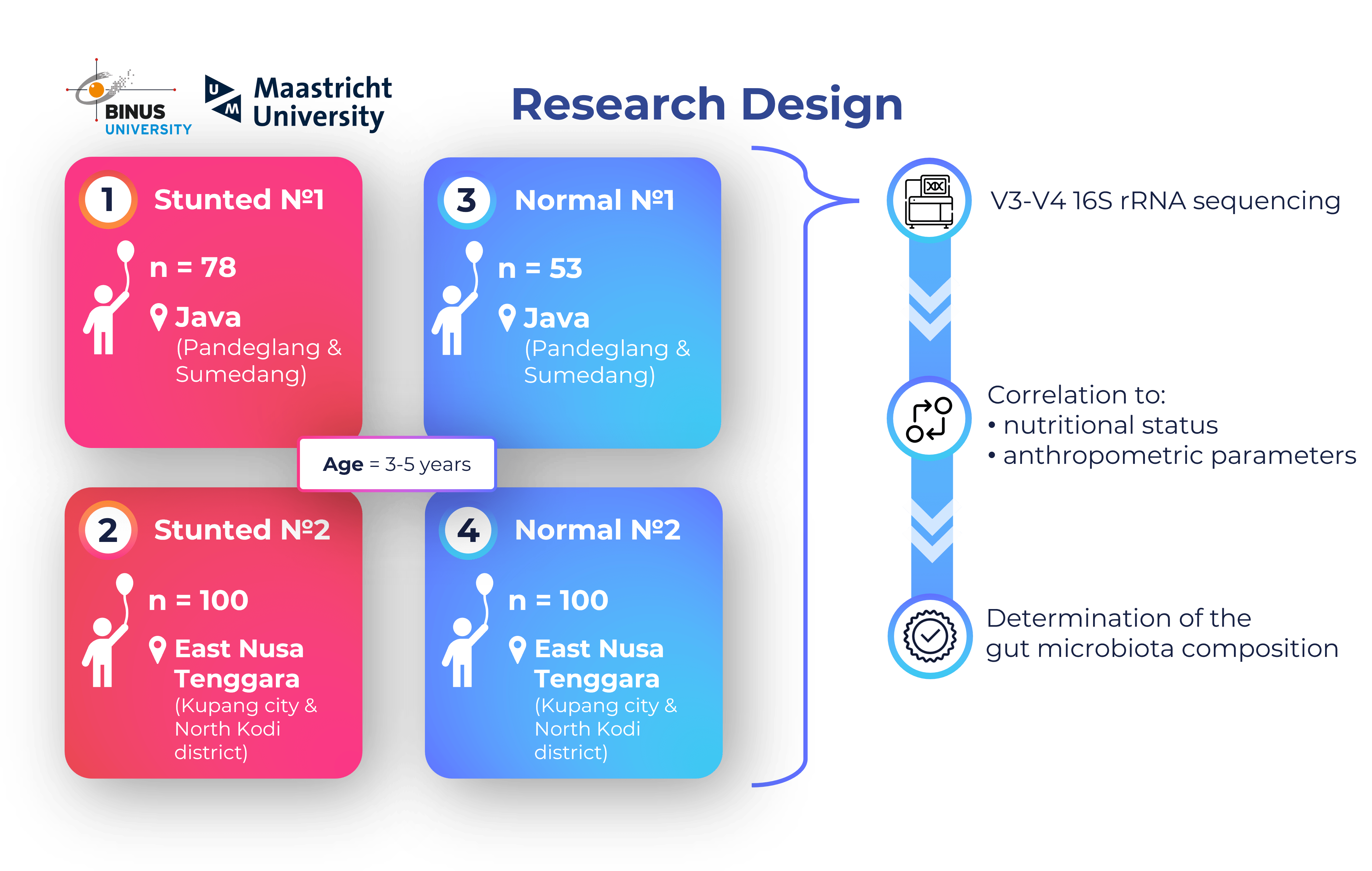
The shadow of HIV
Research Project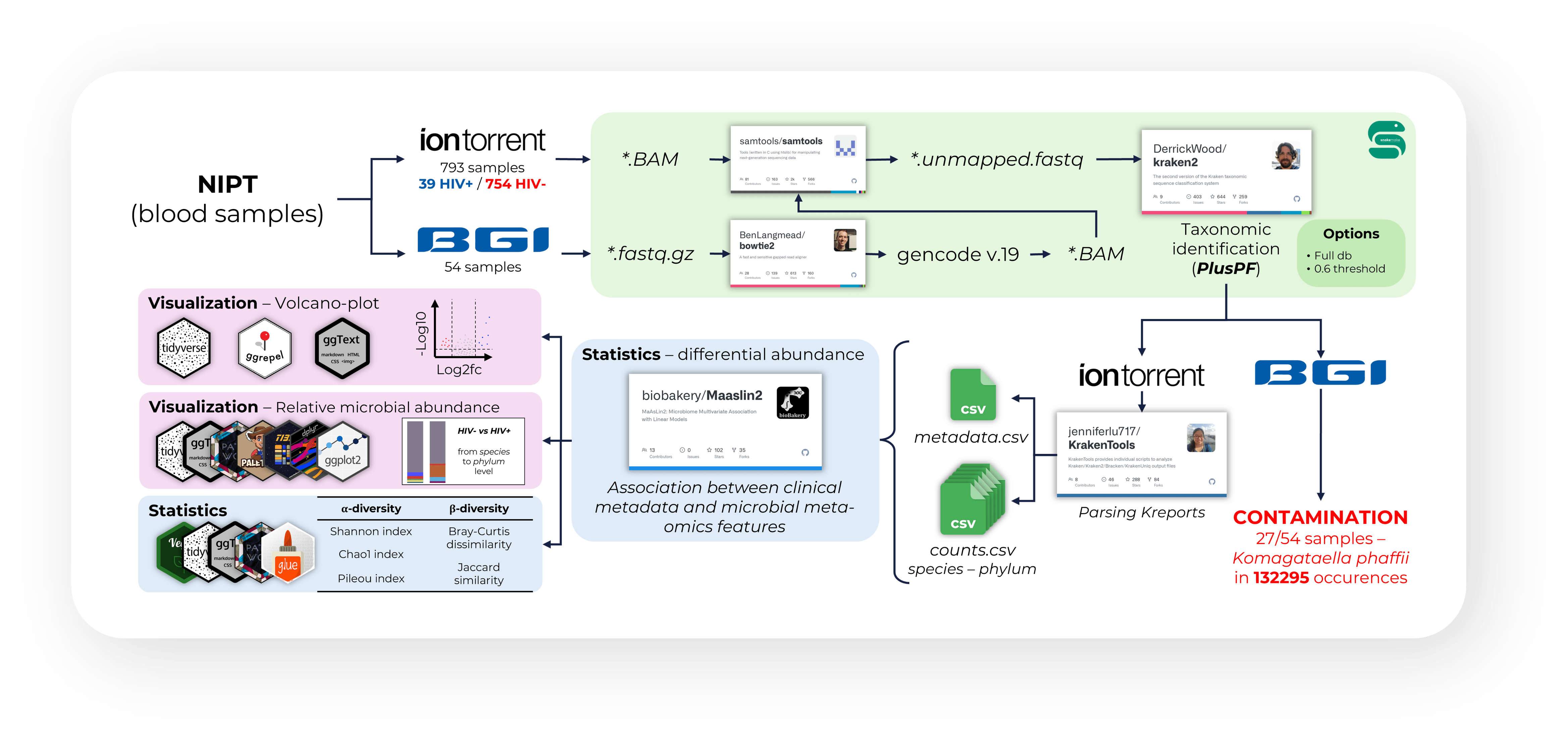
Detection of Brno loanvirus in bats in Southern Russia
Russian Science Foundation project № 23-24-00276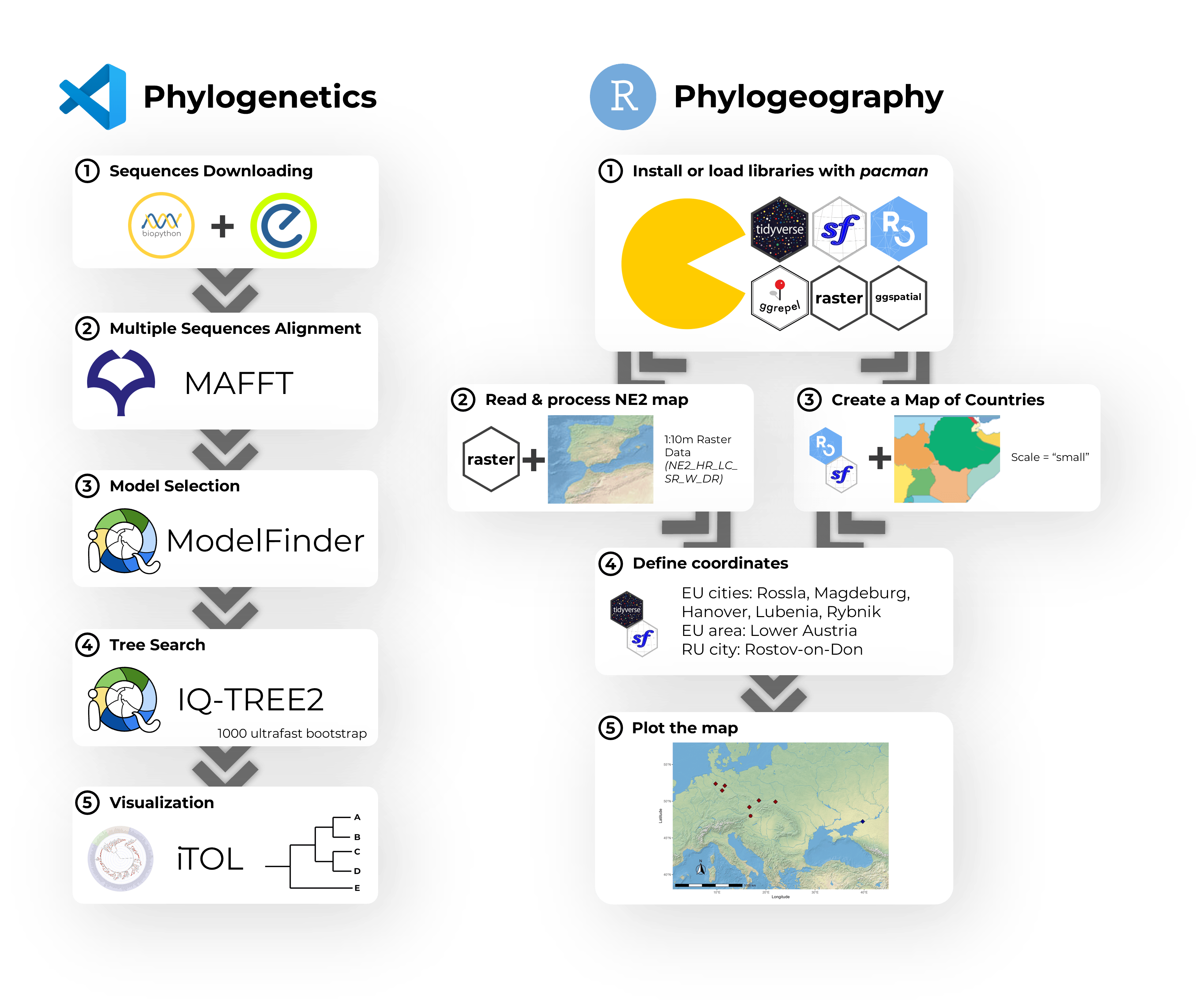
Whole genome metagenomic analysis of the gut microbiome of bats
Russian Science Foundation project № 23-14-00316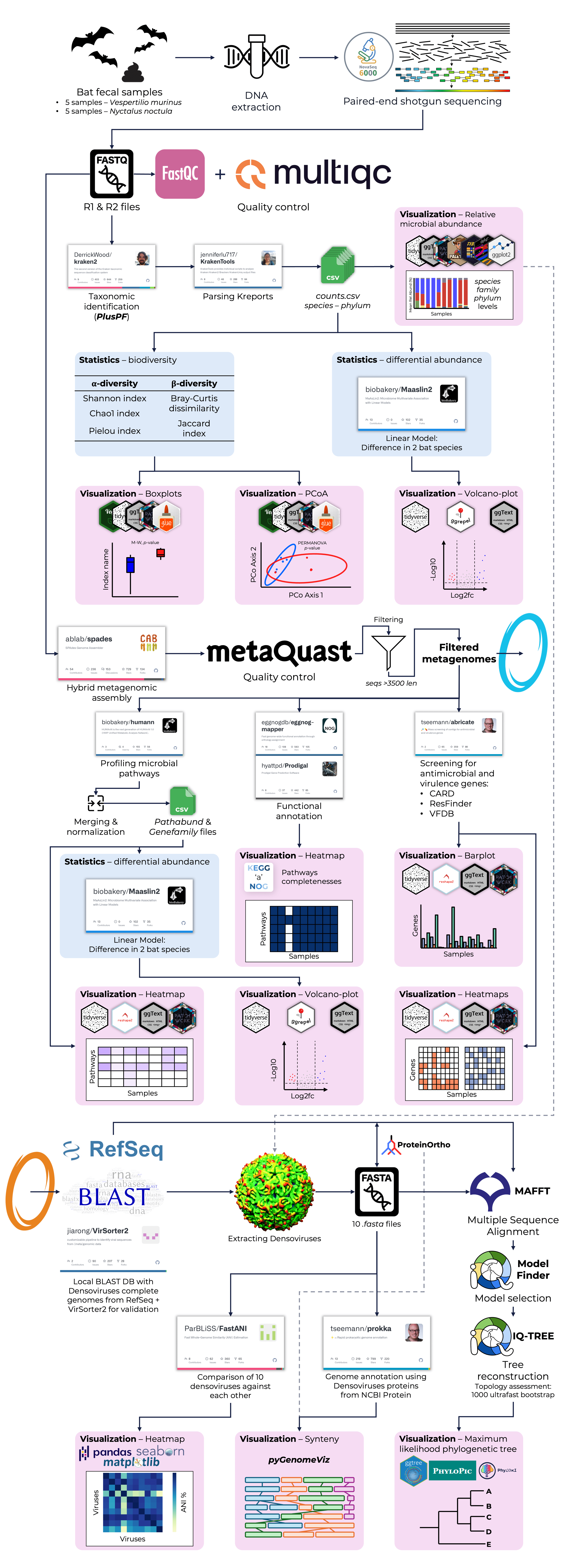
Detection of α-CoV in bats of Rostov and Novosibirsk (Russia)
Russian Science Foundation project № 23-64-00005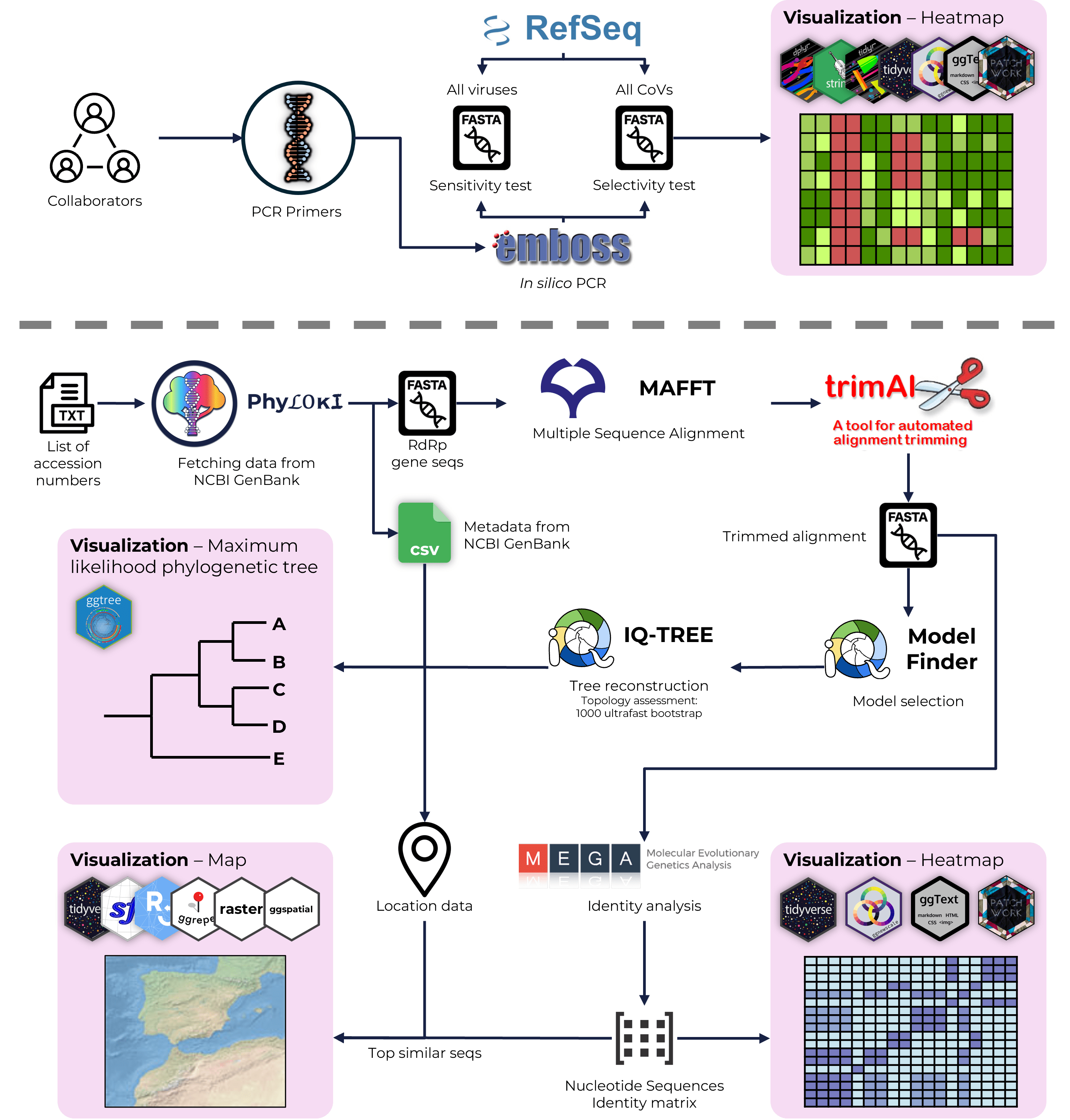
Pan-Salmonella phages degenerate PCR primers design
Russian Science Foundation project № 23-26-00142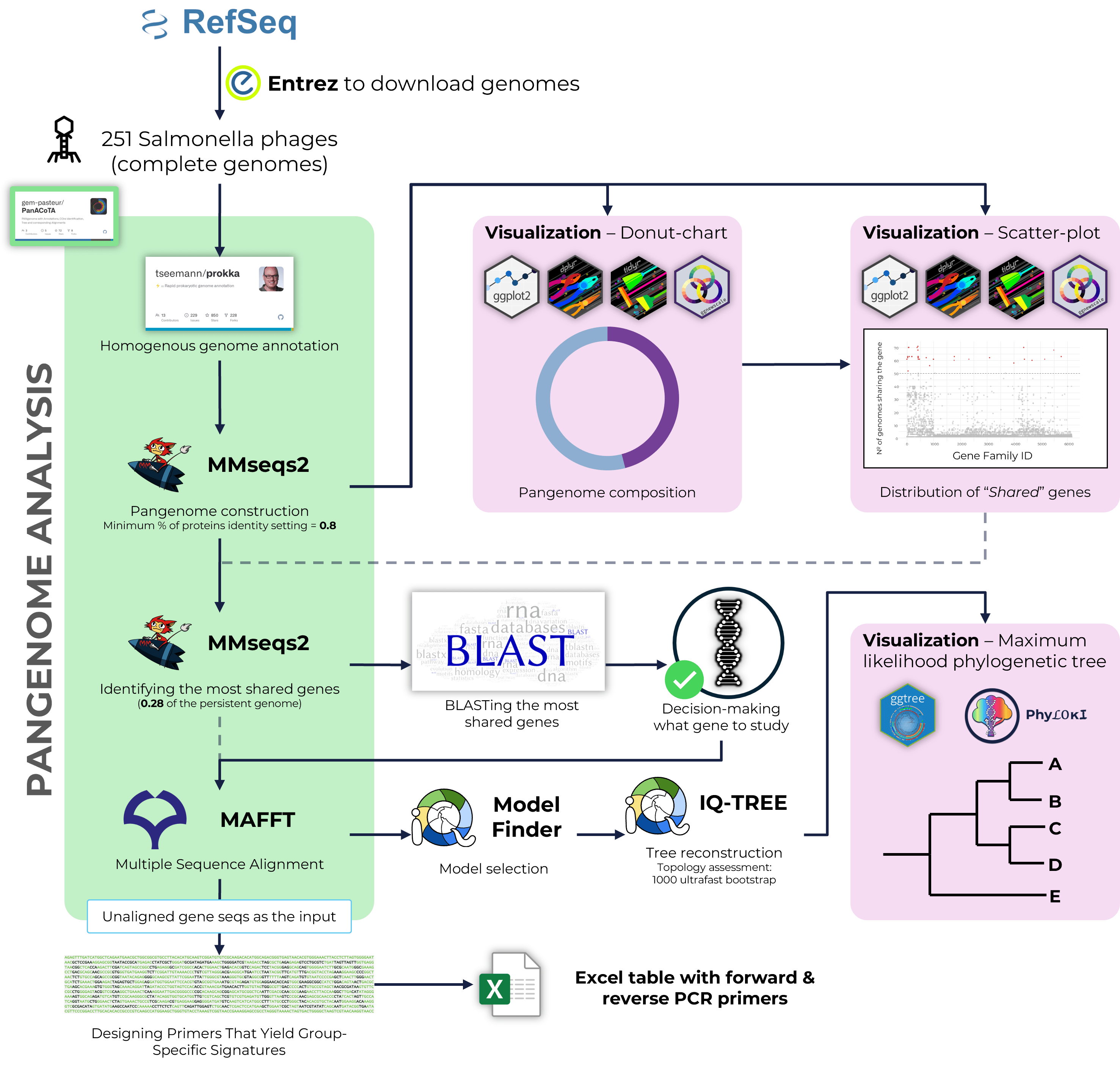
P. destructans: Insights into Dating and Definition Study
Russian Science Foundation project № 25-24-00351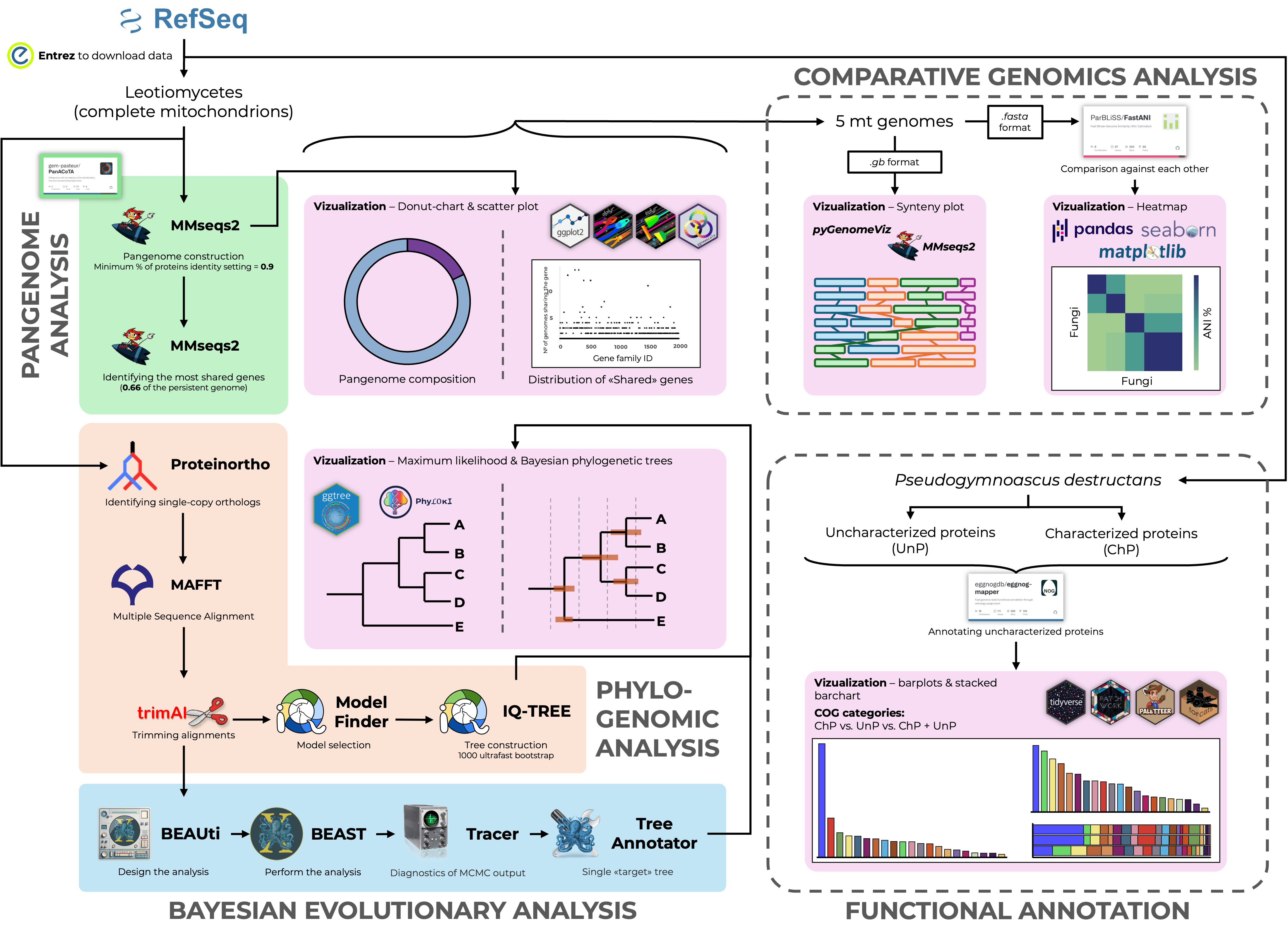
S. albidoflavus SM254 v. P. destructans: Dawn of Justice
Russian Science Foundation project № 25-24-00351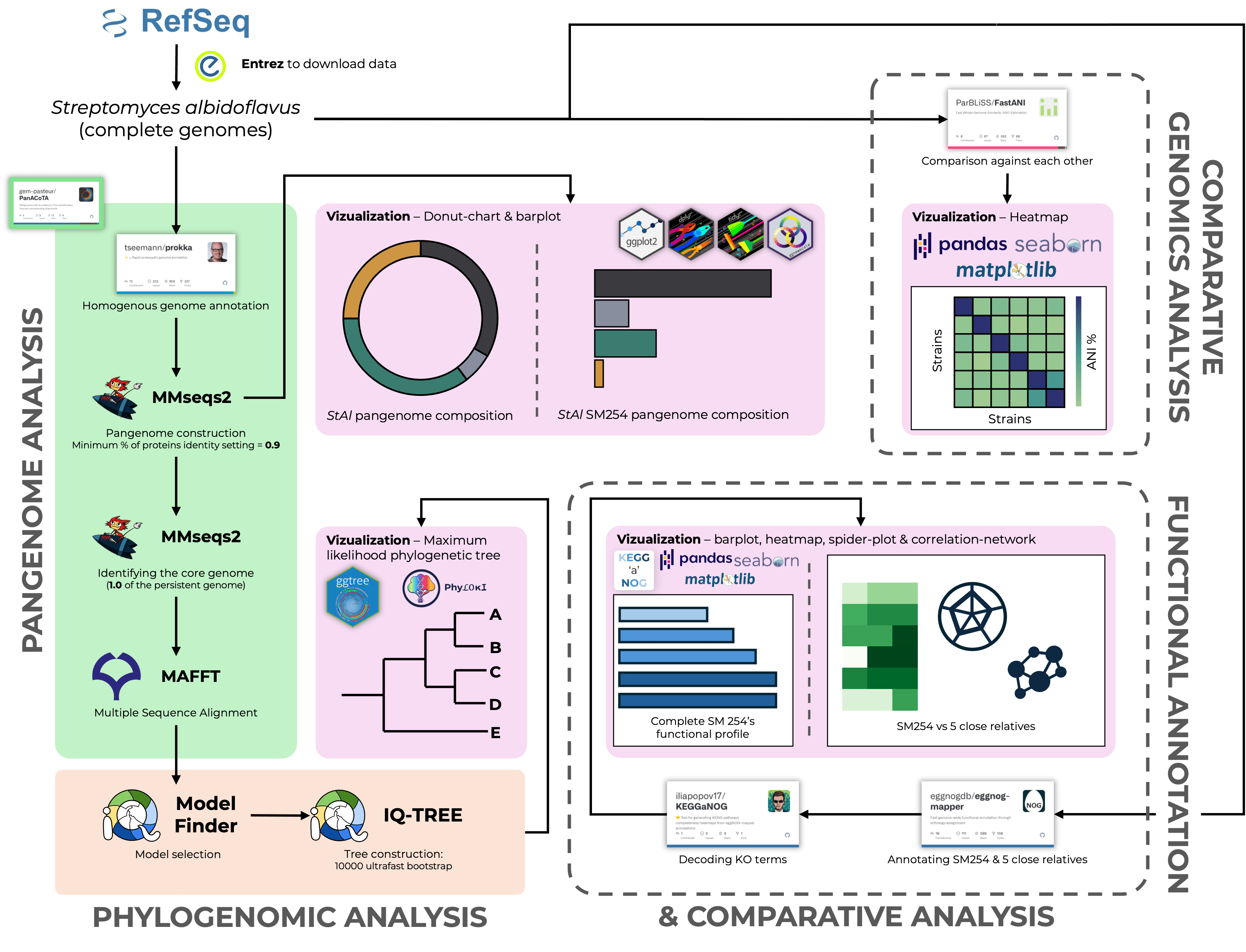
talks
Metagenomic analysis of the intestinal microbiota of synanthropic bats: insect viruses, antibiotic resistance genes and metabolic potential
Web-site / Proceedings / Poster / Certificate
Streptomyces albidoflavus SM254: Genomic markers for counteracting white-nose syndrome in bats
Web-site / Proceedings / Poster / Certificate
teaching
Culture-independent methods for qualitative and quantitative microbiological detection and identification
Lecture for 2nd year undergraduate students who are new to the study of microbiology on «Culture-independent methods for qualitative and quantitative microbiological detection and identification».
NGS Data Analysis
Detailed guidelines on NGS data analysis. Covers topics on: Quality Control of raw data, Genomic Variation Analysis, Whole Genome and Pangenome Analyses, Phylogenetics and 16S Amplicon Analysis.
Introduction to phylogenetic analysis with Python and R programming
A hands-on workshop on phylogenetic analysis using Python and R. The pipeline covered MSA with MAFFT, trimming with trimAl, model selection via ModelFinder, and tree inference using IQ-TREE2. Phyloki were used for metadata integration, with final visualizations generated in ggtree.
tools
MyAwesomeEDA
About: Python module that provides a set of tools for exploring and analyzing your dataset
Status: Released


Phyloki
About: Tool to fetch metadata for phylogenetic trees annotation
Status: Released



KEGGaNOG
About: Tool for generating KEGG heatmaps from eggNOG-mapper annotations
Status: Released & Published




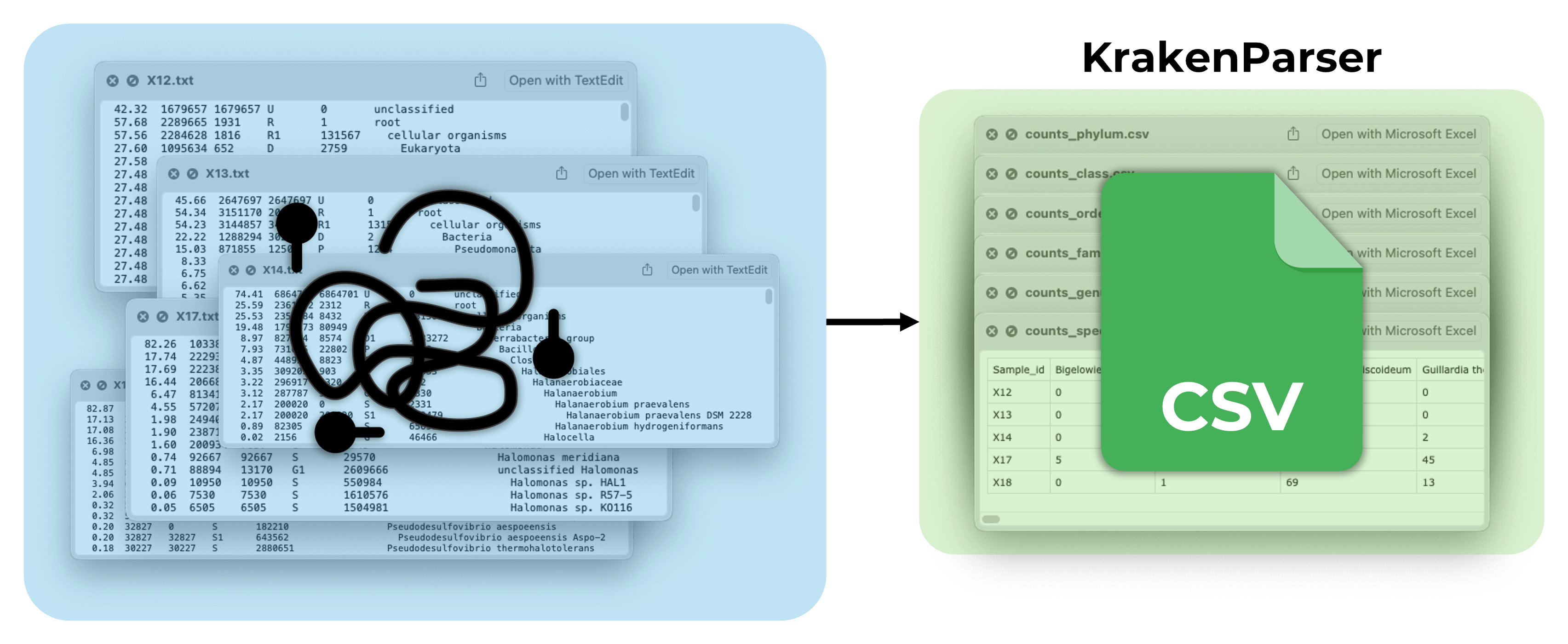
KrakenParser
About: Tool to process Kraken2 reports and convert them into CSV format
Status: Released