Morphogenesis of penile cavernous fibrosis in hypotestosteronemia: an experimental study
Kogan M.I., Todorov S.S., Popov I.V., Popov I.V., Kulishova M.A., Ermakov A.M., Sizyakin D.V.
Urology Herald, 2020
Kogan M.I., Todorov S.S., Popov I.V., Popov I.V., Kulishova M.A., Ermakov A.M., Sizyakin D.V.
Urology Herald, 2020
Kogan M., Popov I., Mitrin B., Popov I., Sadyrin E., Pasetchnik D., Ermakov A., Ugrekhelidze N., Kulikova N.
E3S Web of Conferences, 2020
Donnik I.M., Popov Ig.V., Sereda S.V., Popov Il.V., Chikindas M.L., Ermakov A.M.
Biology Bulletin of the Russian Academy of Sciences, 2021
Kogan M.I., Popov I.V., Kirichenko E.Y., Mitrin B.I., Sadyrin E.V., Kulaeva E.D., Popov I.V., Kulba S.N., Logvinov A.K., Akimenko M.A., Pasechnik D.G., Tkachev S.Yu., Karnaukhov N.S., Lapteva T.O., Sukhar I.A., Maksimov A.Yu., Ermakov A.M.
Andrology, 2021
Sereda T.V., Petrova M.A., Popov I.V., Popov I.V., Kartashov S.N., Ermakov A.M.
Brazilian Journal of Veterinary Medicine, 2022
Lipilkina T., Popov I., Kitsenko K., Popov I., Ermakov A.
E3S Web of Conferences, 2022
Popov I.V., Ohlopkova O.V., Donnik I.M., Zolotukhin P.V., Umanets A., Golovin S.N., Malinovkin A.V., Belanova A.A., Lipilkin P.V., Lipilkina T.A., Popov I.V., Logvinov A.K., Dubovitsky N.A., Stolbunova K.A., Sobolev I.A., Alekseev A.Yu., Shestopalov A.M., Burkova V.N., Chikindas M.L., Venema K., Ermakov A.M.
Scientific Reports, 2023
Popov I.V., Berezinskaia I.S., Popov I.V., Martiusheva I.B., Tkacheva E.V., Gorobets V.E., Tikhmeneva I.A., Aleshukina A.V., Tverdokhlebova T.I., Chikindas M.L., Venema K., Ermakov A.M.
Animals, 2023
Popov I.V., Popov I.V., Krikunova A.A., Lipilkina T.A., Derezina T.N., Chikindas M.L., Venema K., Ermakov A.M.
International Journal of Molecular Sciences, 2023
Surono I.S., Popov I., Verbruggen S., Verhoeven J., Kusumo P.D., Venema K.
PloS ONE, 2024
Popov I.V., Skripkin V.S., Mazanko M.S., Epimakhova E.E., Prazdnova E.V., Dilekova O.V., Dannikov S.P., Popov I.V., Trukhachev V.I., Rastovarov E.I., Derezina T.N., Kochetkova N.A., Weeks R.M., Ermakov A.M., Chikindas M.L.
Journal of Applied Poultry Research, 2024
Ohlopkova O.V., Stolbunova K.A., Popov I.V., Popov I.V., Kabwe E., Davidyuk Yu.N., Stepanyuk M.A., Moshkin A.D., Kononova Yu.V., Lukbanova E.A., Ermakov A.M., Chikindas M.L., Sobolev I.A., Khaiboullina S.F., Shestopalov A.M.
Brazilian Journal of Microbiology, 2024
Ohlopkova, O.V., Popov, I.V., Popov, I.V., Stolbunova, K.A., Stepanyuk, M.A., Moshkin, A.D., Maslov, A.A., Sobolev, I.A., Malinovkin, A.V., Tkacheva, E.V., Bogdanova, D.A., Lukbanova, E.A., Ermakov, A.M., Alekseev, A.A., Todorov, S.D., Shestopalov, A.M.
Microbiology Research, 2024
Popov I.V., Popov I.V., Chebotareva I.P., Tikhmeneva I.A., Peshkova D.A., Krikunova A.A., Tkacheva E.V., Algburi A.R., Abdulhameed A.M., Jargalsaikhan A., Ganbold O., Chikindas M.L., Venema K., Ermakov A.M.
Brazilian Journal of Microbiology, 2025
Semizhon P.A., Scheslenok E.P., Dubkov N.A., Sukhotskaya E.A., Stolbunova K.A., Popov I.V., Popov I.V., Alekseev A.Y., Kabwe E., Davidyuk Y.N.
Problems of Virology, 2025
Popov I.V., Peshkova D.A., Lukbanova E.A., Tsurkova I.S., Emelyantsev S.A., Krikunova A.A., Malinovkin A.V., Chikindas M.L., Ermakov A.M., Popov I.V.
Veterinary Sciences, 2025
Popov I.V., Manakhov A.D., Gorobets V.E., Diakova K.B., Lukbanova E.A., Malinovkin A.V., Venema K., Ermakov A.M., Popov I.V.
International Journal of Molecular Sciences, 2025
Popov I.V., Chikindas M.L., Popov I.V.
Brazilian Journal of Microbiology, 2025
Popov I.V., Todorov S.D., Chikindas M.L., Venema K., Ermakov A.M., Popov I.V.
Journal of Fungi, 2025
Lipilkina T.A., Alves M.V., Carneiro K.O., Leani K., Popov I.V., Popov I.V., Chikindas M.L., Ermakov A.M., Todorov S.D.
Molecular Nutrition & Food Research, 2025
Popov I.V., Chikindas M.L., Venema K., Ermakov A.M., Popov I.V.
Molecular Nutrition & Food Research, 2025
Web-site / Proceedings / Poster / Certificate
Web-site / Proceedings / Poster / Certificate
Lecture for 2nd year undergraduate students who are new to the study of microbiology on «Culture-independent methods for qualitative and quantitative microbiological detection and identification».
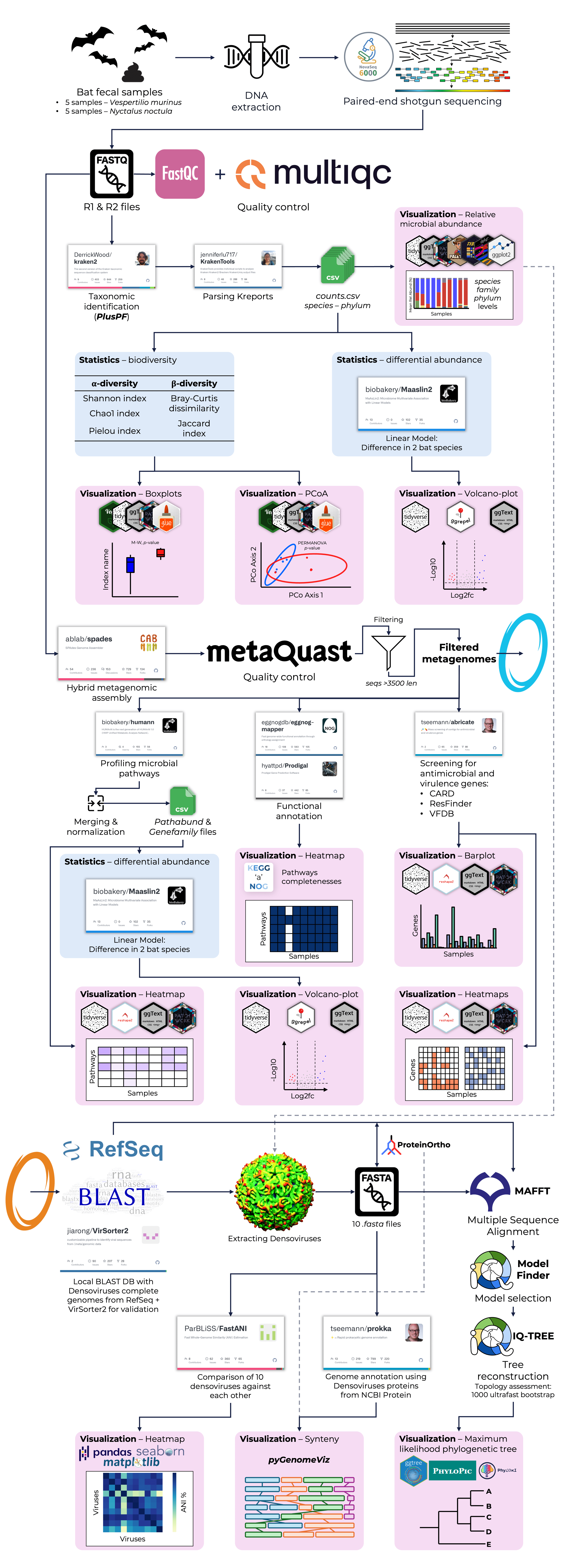
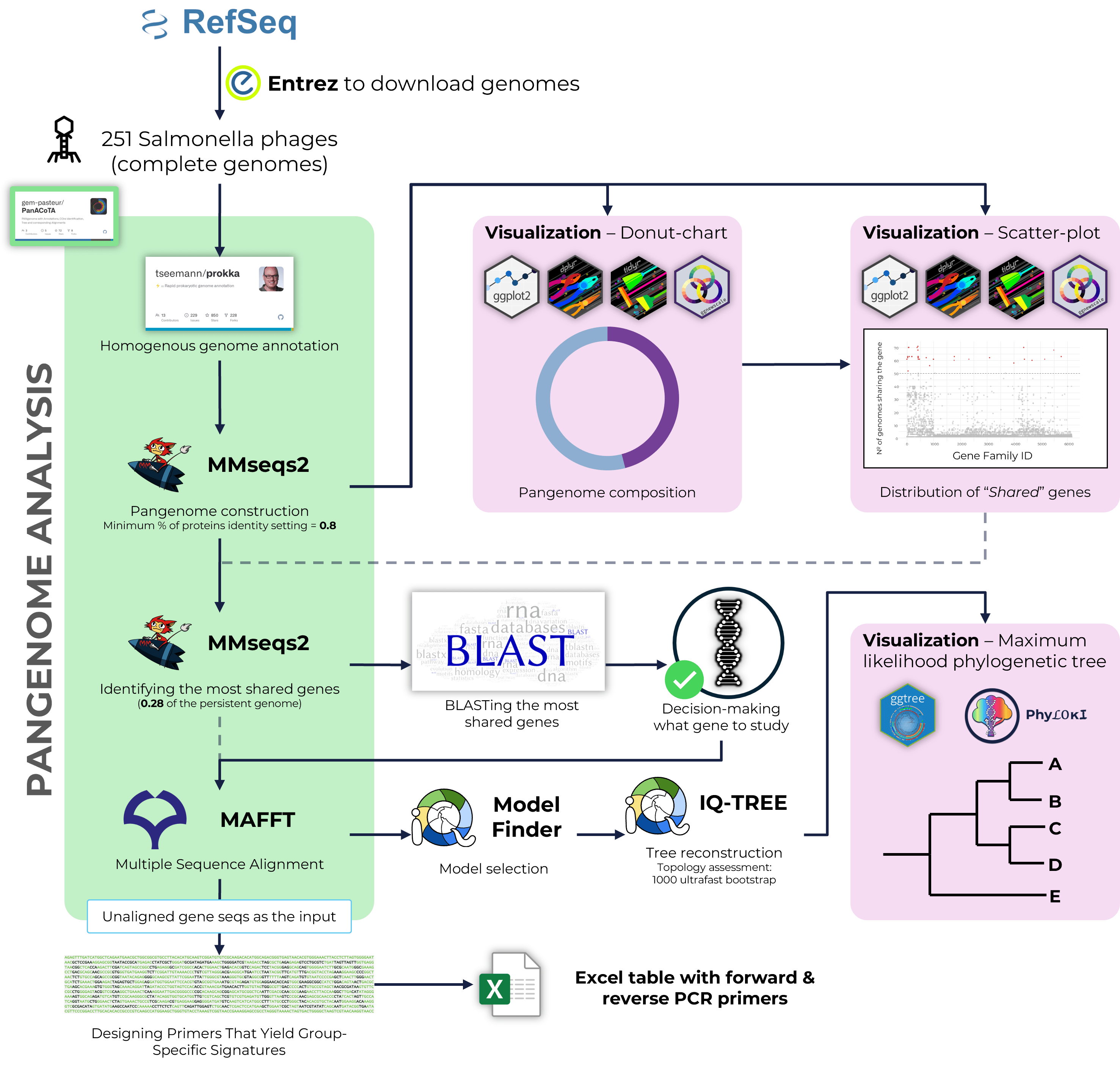
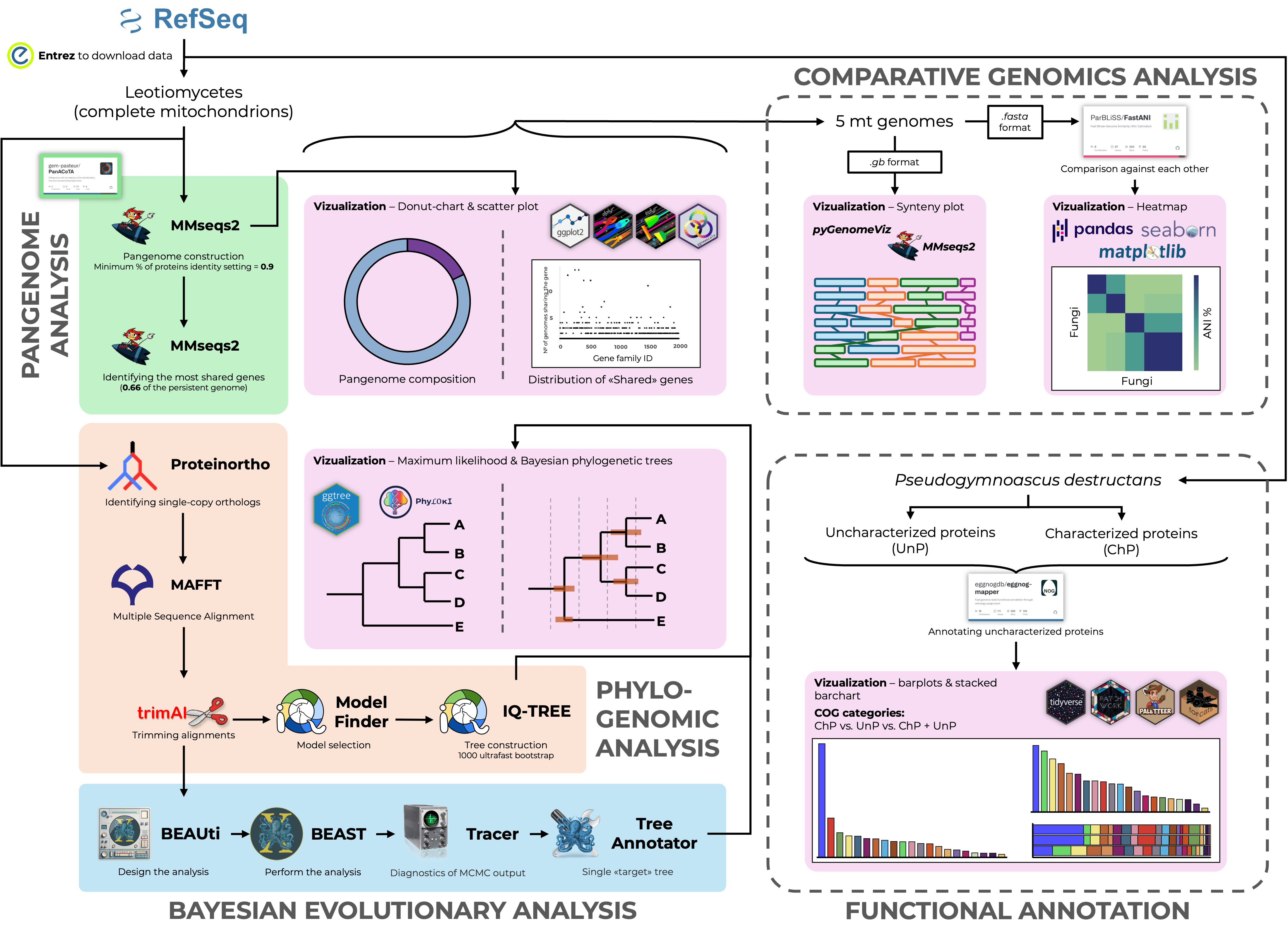
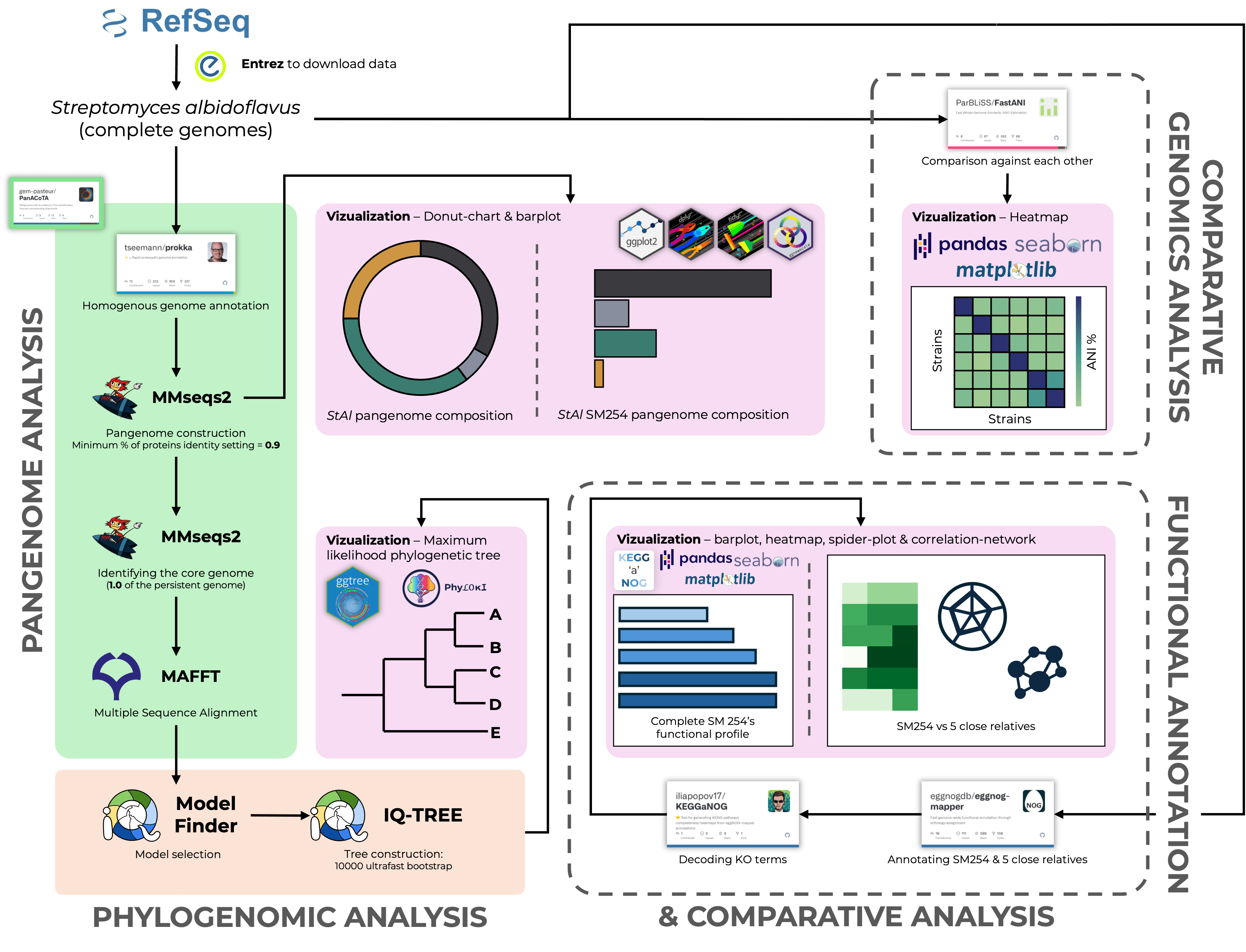
Detailed guidelines on NGS data analysis. Covers topics on: Quality Control of raw data, Genomic Variation Analysis, Whole Genome and Pangenome Analyses, Phylogenetics and 16S Amplicon Analysis.
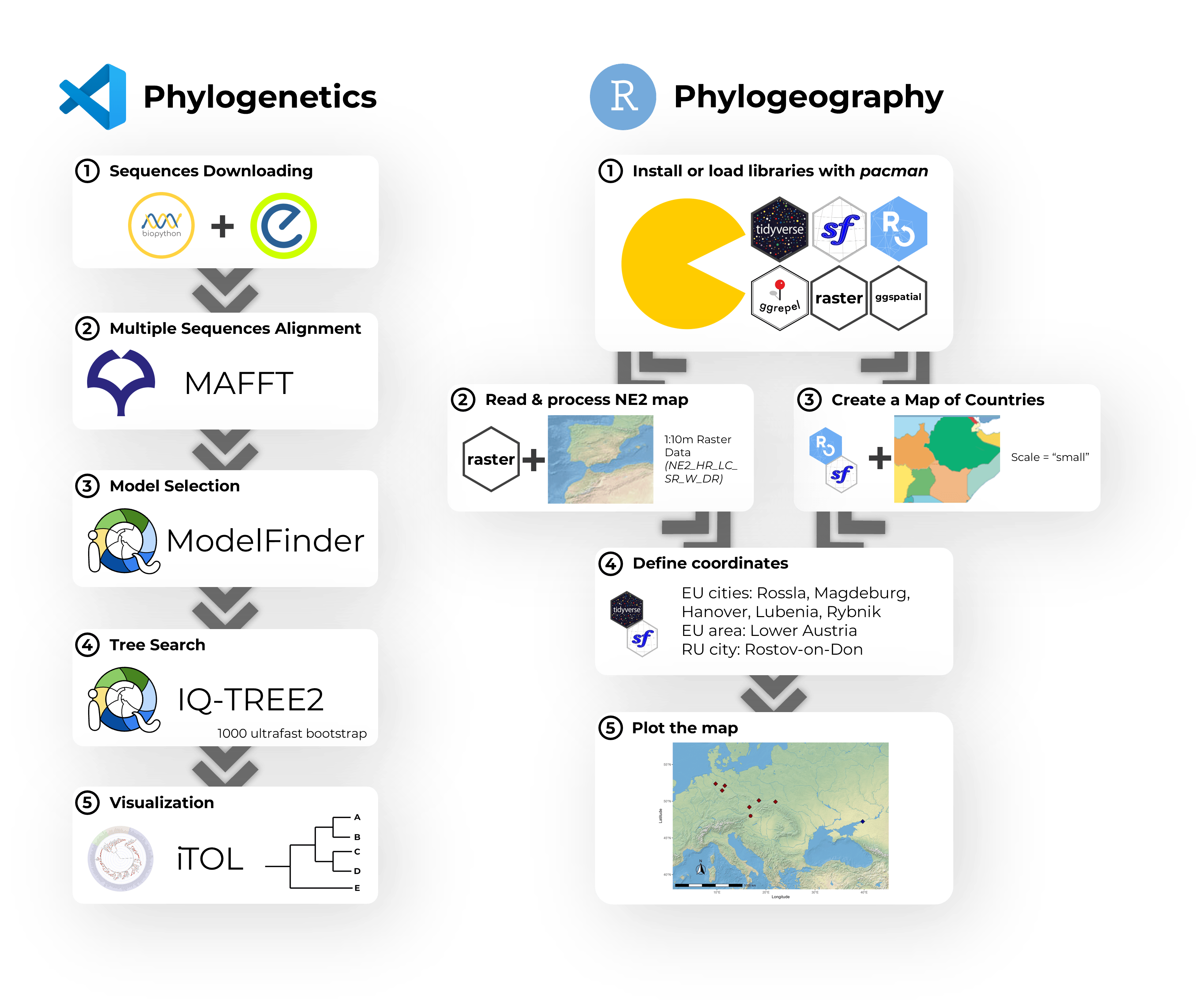
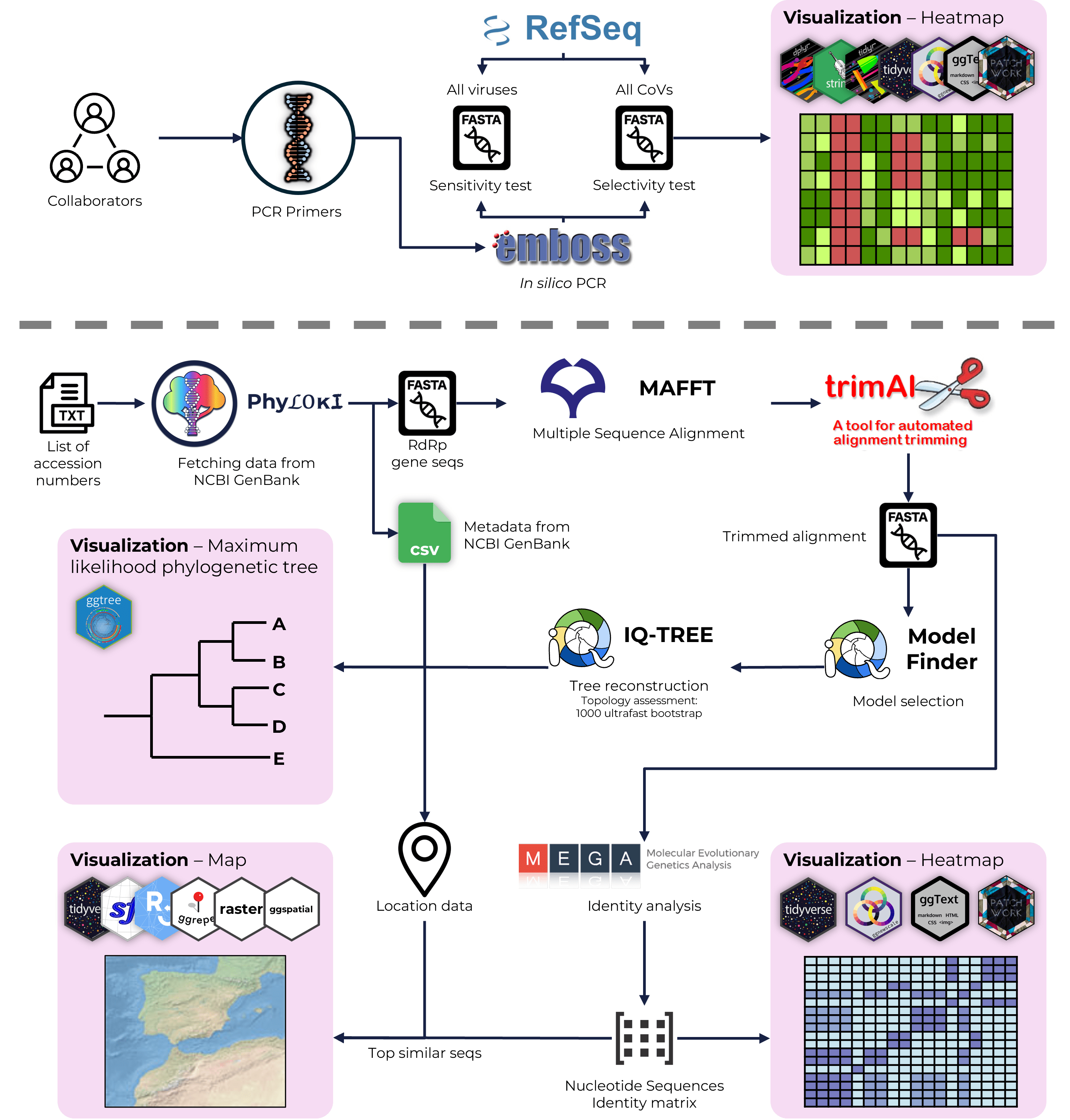
A hands-on workshop on phylogenetic analysis using Python and R. The pipeline covered MSA with MAFFT, trimming with trimAl, model selection via ModelFinder, and tree inference using IQ-TREE2. Phyloki were used for metadata integration, with final visualizations generated in ggtree.
About: Python module that provides a set of tools for exploring and analyzing your dataset
Status: Released


About: Tool to fetch metadata for phylogenetic trees annotation
Status: Released



About: Tool for generating KEGG heatmaps from eggNOG-mapper annotations
Status: Released & Published




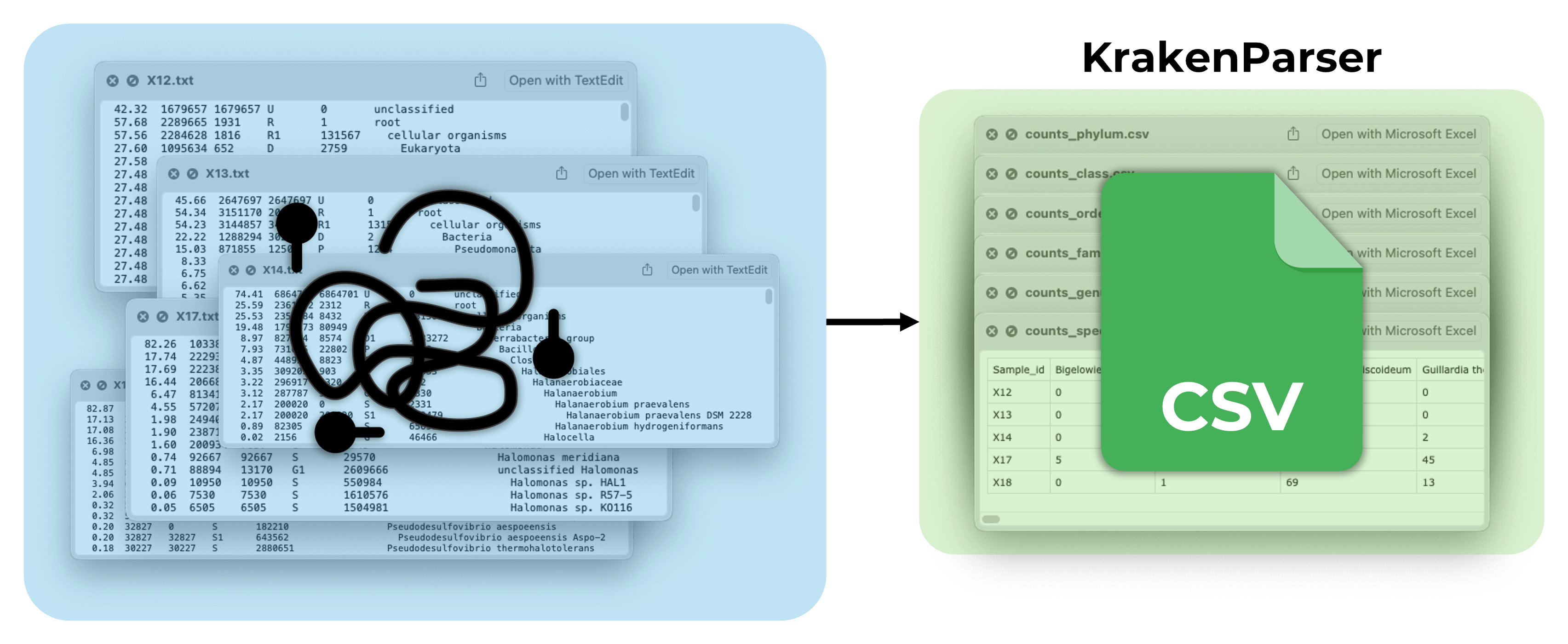
About: Tool to process Kraken2 reports and convert them into CSV format
Status: Released