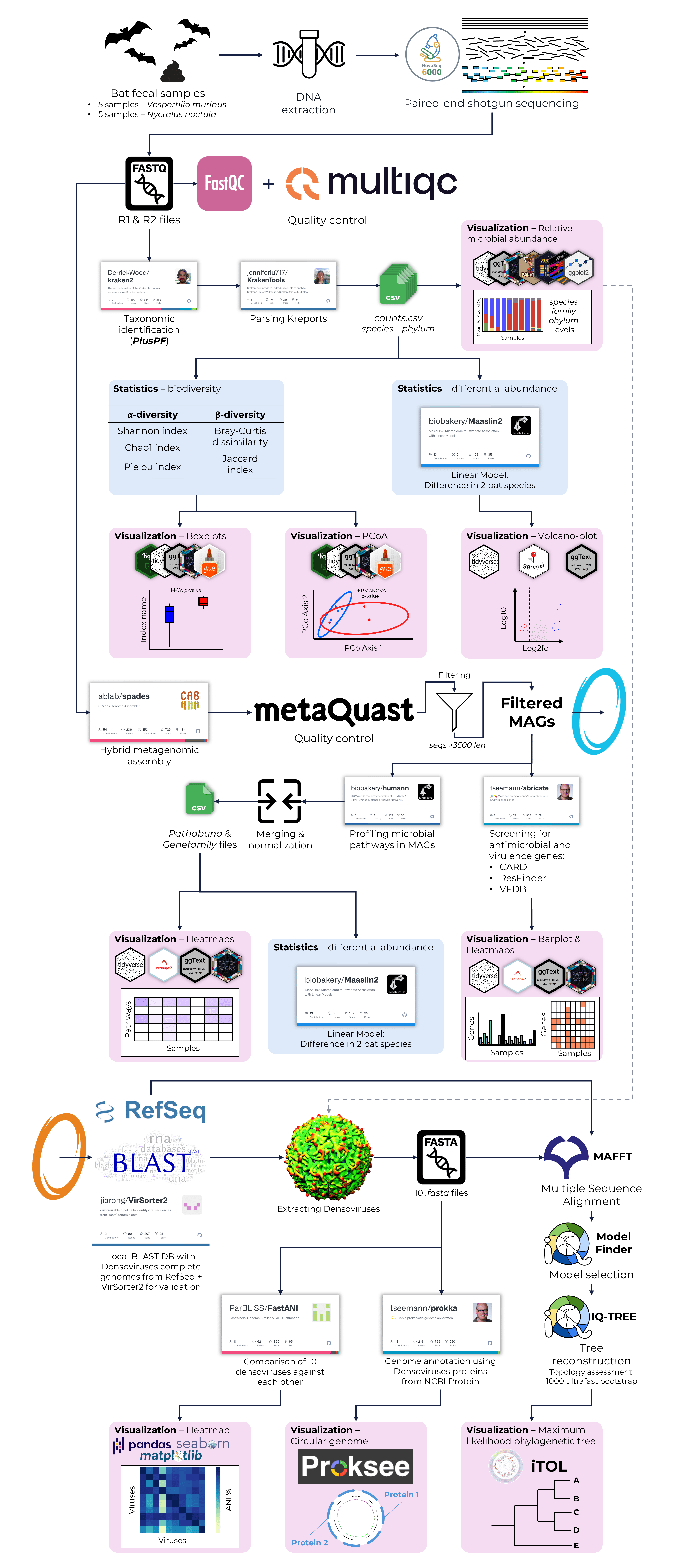
Pediatric Nutritional Medicine and Intestinal Microbiology
Research Internship in Indonesia (Bina Nusantara University; Feb 2023 – July 2023)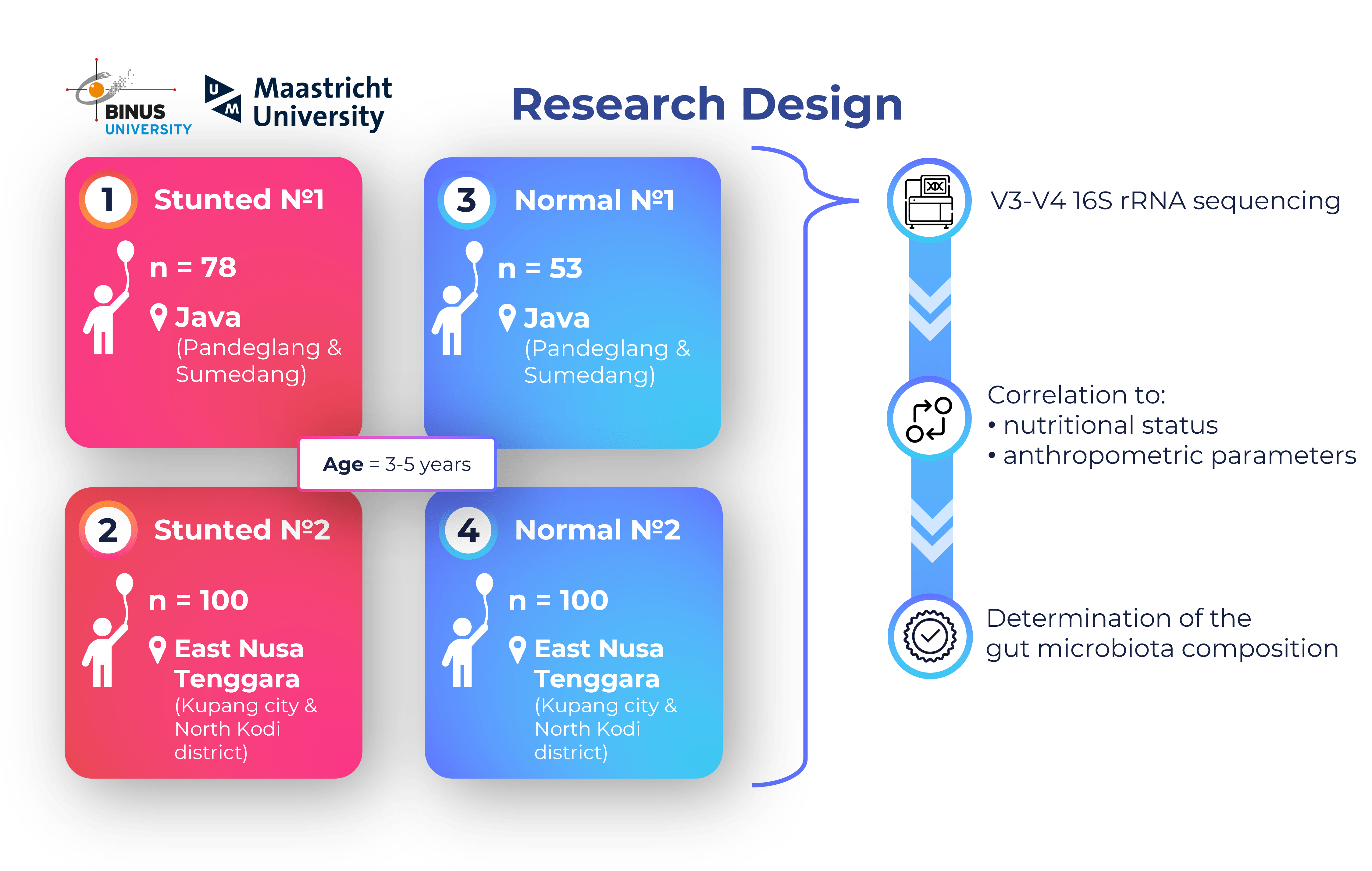
Research Internship in Indonesia (Bina Nusantara University; Feb 2023 – July 2023)
Research Project (Bioinformatics Institute; Feb 2024 – May 2024)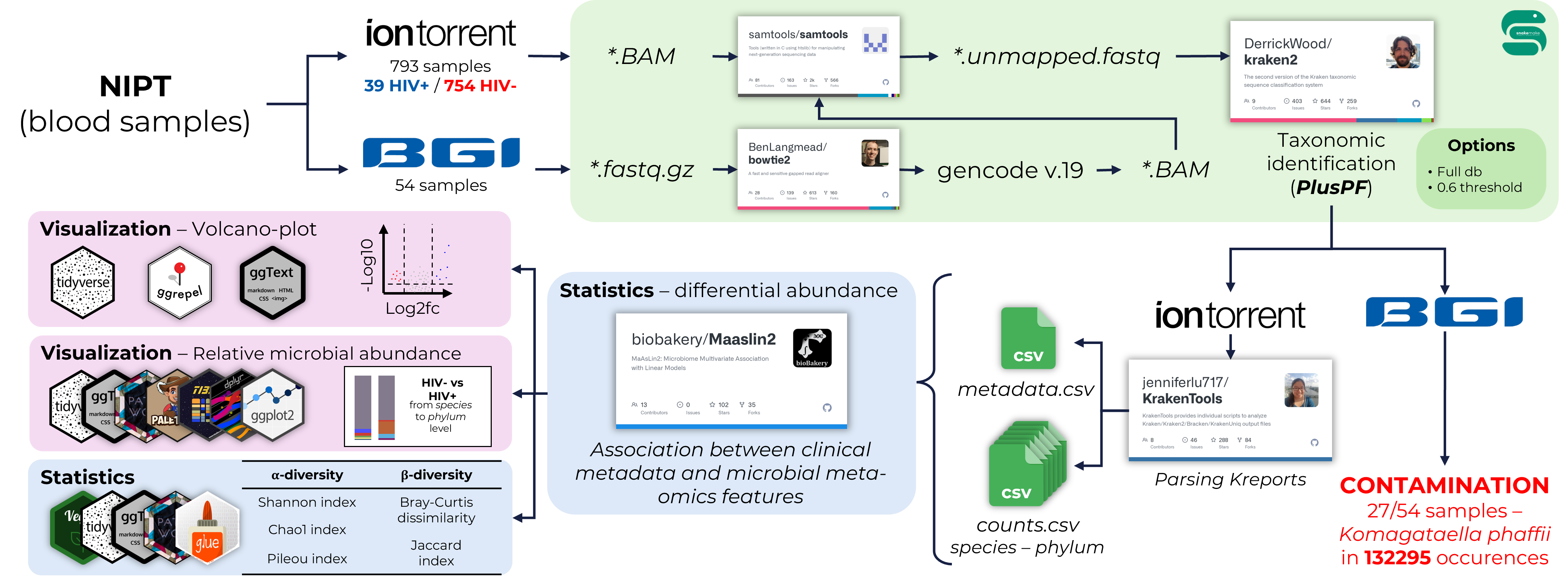
Study projects completed as part of the workshops undertaken during the training at the Bioinformatics Institute:
1. Variant calling of Escherichia coli WGS
2. Variant calling of deep sequencing data (Influenza A virus (H3N2) hemagglutinin gene)
3. De novo assembly of Escherichia coli genome
4. Tardigrade Ramazzottius varieornatus genome annotation and protein function prediction
5. Genotyping and SNP annotation of human 23andMe data
6. RNA-seq data analysis for differential gene expression of Saccharomyces cerevisiae after 30 minutes of fermentation
7. Ancient metagenomes analysis examining human dental calculus
8. Annotation of the immune repertoire derived from the T-cell population in a relatively healthy donor
9. Single-cell CITE-seq analysis detailing the cellular composition and transcriptional profiles within human bone marrow
Russian Science Foundation grant research project (Don State Technical University; May 2024)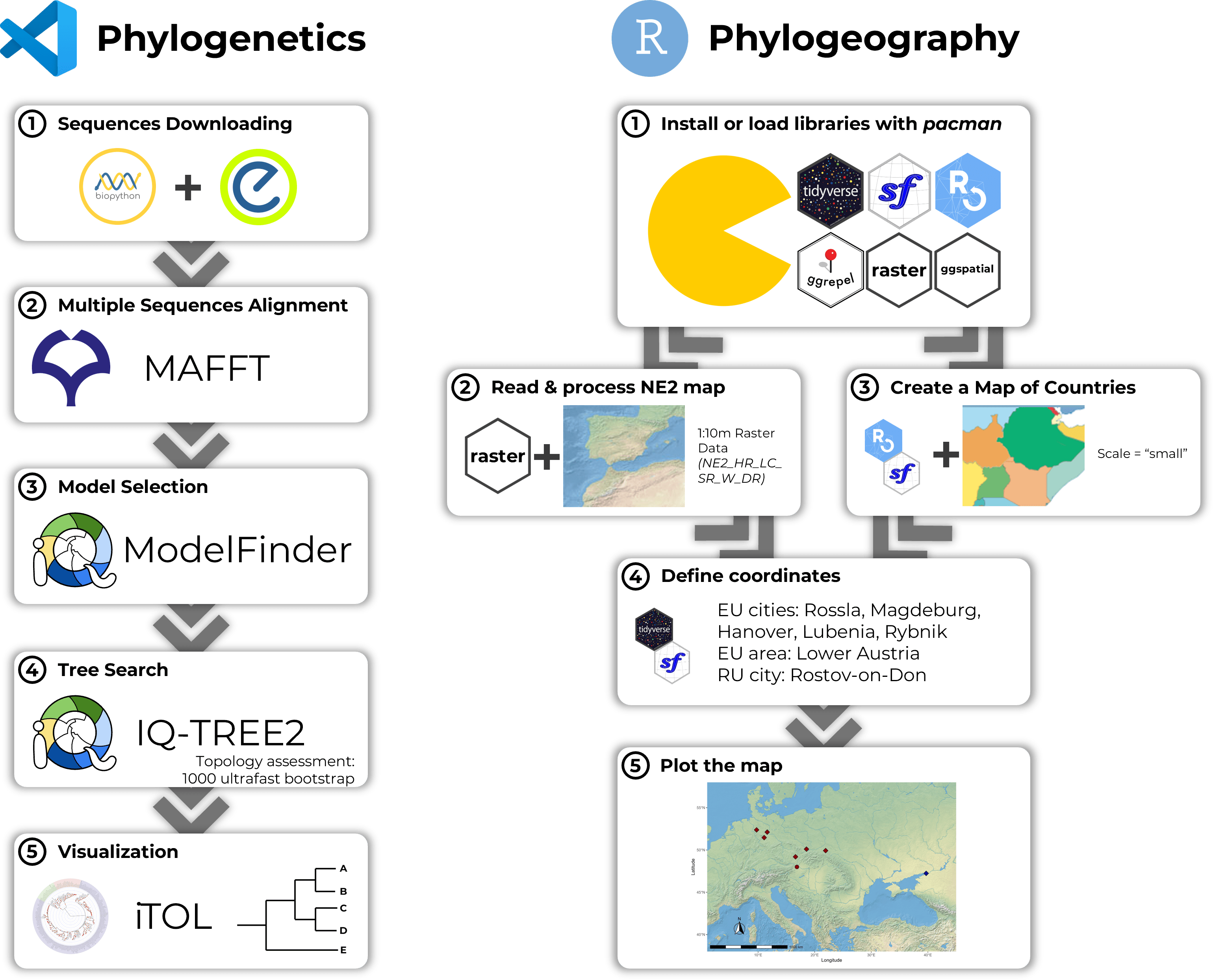
Russian Science Foundation grant research project (Don State Technical University; June 2024 – Aug 2024)